Meetups are now bimonthly, at the Hacklab.
October 19th, 2025
This coming Sunday, we will be hosting an Intro to R course, similar to the one last spring. This is the first in a series of workshops on R programming in Biology.
Saturday, October 4th, 2025
We hosted a fun and informative PCR Workshop at Deer Park Library, near St. Clair subway station. Participants learnt to pipette, perform PCR and load an agarose gel!
Dec. 20, 2024
As our planet spins around the sun, and we are almost turning into a new year (Solstice is tmro!), it’s time to take stock of the past year, and all the good stuff that has happened at DIYBIO.to this year.
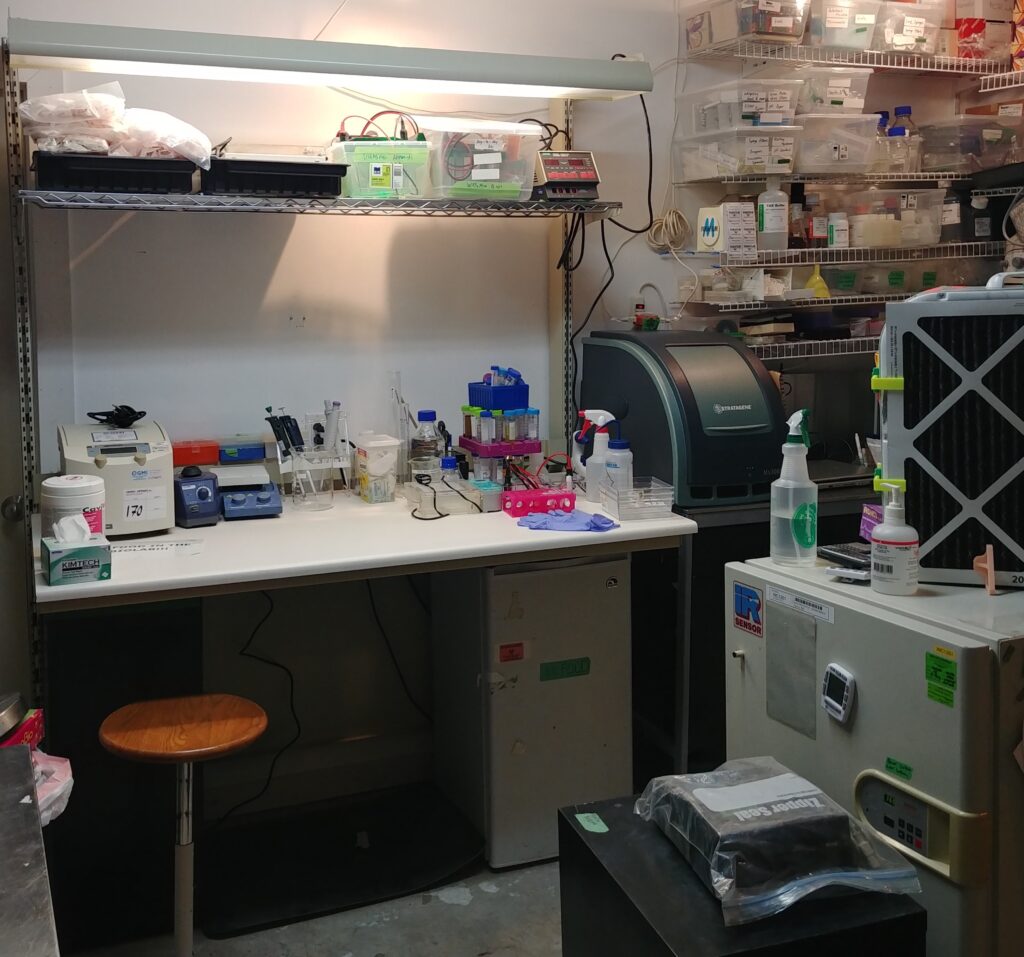
Donations and acquisitions news !!!!
An amazing amount of stuff has been donated over the last year from friends and members of DIYBIO.
A huge shout out to Aaron, Julie, Rana and Norm for donating over the last year!!!
We now have a better microfuge with controllable speed, protein minigel apparati for protein gels, and consumables for regular lab work. I also provided a new bench where I hope we can be super productive!
Bioinformatics group news
Heraclitis is stated something like “Noone ever steps in the same river twice”, meaning that change is the only constant. This is nowhere more true than in bioinformatics. We are interested in providing some basic training and discussion in this area, as it is important for early stage science students to get a grasp of, but also for the public to be able to access and understand data of their own, or public data. This information is freely available, and as a community group that is interested in encouraging public engagement in biology and computers, we have meetups to encourage this. But to get through this we need to define our objectives clearly.
Some of us at DIYBIO are interested in making bioinformatics for accessible using: 1. gene expression in datasets that are publicly available and 2. metabolic pathway changes in non-health related data (plant for instance) as an example of how any genetic expression data can be used.
It is important to note that the source of the original data in health bioinformatics can influence how we interpret results. For instance, background data on gene ontology and pathways can preferentially weight oncologic data, due to the vast amount of data in that area being added to KEGG, webgestalt, PANTHER, DAVID and other sites. Even if analysis is weighted and an appropriate reference dataset is used, there is just so much more historical data in the cancer area that cancer pathways will likely be enriched and found in non-cancer disease. This can confound the ability to find unique pathways for unique understudied diseases. It can also potentially weight cancer studies toward the most studied cancers. For this reason, a more historical-data agnostic approach may be needed, such as a transcriptional approach. For instance, looking at the potential binding partners in upregulated genes, would be agnostic to historical pathway data on cancer. For example, cytohubba and iRegulon in R and Cytoscape, DIRE and Opossum could be used, which is what we are presently doing!
In other bioinformatics approaches, we are looking at the absolute latest in advances in high resolution spatial genomics and trying to tease apart the potential approaches in these new techniques. The data available online is astounding! But using it in downstream methods is relatively new and challenging. Despite this, we have already performed some initial analysis of public datasets using pseudobulk methods after running Seurat and SpaCET, then DESeq2. Ask at our weekly meetups, diybio email or the hacklab open house, if you are interested in contributing to this project.
Nov. 27, 2024

Offsite trip to CRAFT at U of T!
So much cool tech! We saw:
- wearable technology production with the Roll-2-Roll Polymer Coater!
- laser cutters! Multiple microchip printers that work with plastic, glass or resin!
- Many more machines including the piece de resistance, the Injection molder!
Lots of discussion about what these are used for, and how they could be useful for member projects.
Cheers!
- Madlab